Chromosome Structure and Gene Expression
The chromosomes of multicellular animals have a regular and inheritable physical organization. This was first recognized in studies dating back to the 1890’s on the lampbrush chromosomes in amphibian oocytes (Fig.1) and the polytene chromosomes of insects. Subsequent studies have shown that the architectural principles inferred from analysis of lampbrush and polytene chromosomes are universal features of chromosomes throughout much of the animal kingdom. The key organizational principle is the subdivision of the chromatin fiber into a series of independent looped domains, called “TADs.” The arrangement of TADs along the chromosome tend to be invariant and are largely independent of the cell type or developmental stage. This regular and inheritable organization is a reflection of the underlying mechanism of TAD formation. TADs are separated from each other by special elements called boundaries or insulators. While these elements have been found in many different species, they have been most fully characterized in Drosophila. Fly boundaries span DNA sequences of 150 bp to 1.5 kb in length and contain one or more nucleosome-free nuclease-hypersensitive regions. These nuclease-hypersensitive regions are targets for a large collection of DNA binding proteins that have been implicated in boundary function.
Boundary elements in flies are not only responsible for organizing the chromatin fiber, they also have genetic activities. When interposed between enhancers or silencers and target promoters, boundary elements block regulatory interactions This insulating activity provides a mechanism for delimiting units of independent gene activity: genes located between a pair of compatible boundaries are subject to regulatory interactions with enhancers/silencers present in the same chromosomal interval, while they are insulated from the effects of enhancers/silencers located beyond either boundary in adjacent regulatory neighborhoods. Genetic studies suggest that the insulating activity of boundary elements is a consequence of subdividing the chromosome into a series of topologically independent domains. Organizing the chromatin fiber into looped domains enhances contacts between sequences within the loop, while it suppresses contacts with sequences outside of the loop.
Our current research efforts are aimed at determining how boundaries function. A combination of genetic and molecular studies indicate that fly boundaries are functionally non-autonomous and that their activities in both loop formation and gene regulation depend upon their ability to engage in direct physical interactions with other boundaries. Physical interactions are mediated by the DNA binding proteins associated with potential pairing partners. Many fly chromosomal architectural proteins (Pita, Zipic, CTCF) form homomultimers. Consequently, if two boundaries share binding sites for one or more of these proteins, the two boundaries can be linked together by the multimer. In other cases, two different proteins can link two boundaries together by forming heteromeric complexes.
Boundary pairing interactions have several important properties which we are investigating. These include partner preferences and orientation dependence. The properties have important implications both for the formation of TADs, as well as TADs within TADs, and for long distance regulatory interactions.
Selected Publications:
1) The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. Udvardy A, Maine E, Schedl P.J Mol Biol. 1985 Sep 20;185(2):341-58. doi: 10.1016/0022-2836(85)90408-5.PMID: 2997449
2) A position-effect assay for boundaries of higher order chromosomal domains. Kellum R, Schedl P. Cell. 1991 Mar 8;64(5):941-50. doi: 10.1016/0092-8674(91)90318-s.PMID: 1848159
3) A group of scs elements function as domain boundaries in an enhancer-blocking assay. Kellum R, Schedl P.Mol Cell Biol. 1992 May;12(5):2424-31. doi: 10.1128/mcb.12.5.2424-2431.1992.PMID: 1569958
4) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Hagstrom K, Muller M, Schedl P.Genes Dev. 1996 Dec 15;10(24):3202-15. doi: 10.1101/gad.10.24.3202.PMID: 8985188
5) The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P.Genetics. 1999 Nov;153(3):1333-56. doi: 10.1093/genetics/153.3.1333.PMID: 10545463
6) Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Vazquez J, Schedl P.Genetics. 2000 Jul;155(3):1297-311. doi: 10.1093/genetics/155.3.1297.PMID: 10880489
7) Protein:protein interactions and the pairing of boundary elements in vivo. Blanton J, Gaszner M, Schedl P.Genes Dev. 2003 Mar 1;17(5):664-75. doi: 10.1101/gad.1052003.PMID: 12629048
8) Mechanism of chromosomal boundary action: roadblock, sink, or loop? Gohl D, Aoki T, Blanton J, Shanower G, Kappes G, Schedl P.Genetics. 2011 Mar;187(3):731-48. doi: 10.1534/genetics.110.123752. Epub 2010 Dec 31.PMID: 21196526
9) Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Aoki T, Sarkeshik A, Yates J, Schedl P.Elife. 2012 Dec 13;1:e00171. doi: 10.7554/eLife.00171.PMID: 23240086
10) Determinants of Chromosome Architecture: Insulator Pairing in cis and in trans. Fujioka M, Mistry H, Schedl P, Jaynes JB.PLoS Genet. 2016 Feb 24;12(2):e1005889. doi: 10.1371/journal.pgen.1005889. eCollection 2016 Feb.PMID: 26910731
11). Functional Dissection of the Blocking and Bypass Activities of the Fab-8 Boundary in the Drosophila Bithorax Complex. Kyrchanova O, Mogila V, Wolle D, Deshpande G, Parshikov A, Cléard F, Karch F, Schedl P, Georgiev P.PLoS Genet. 2016 Jul 18;12(7):e1006188. doi: 10.1371/journal.pgen.1006188. eCollection 2016 Jul.PMID: 27428541
12) The bithorax complex iab-7 Polycomb response element has a novel role in the functioning of the Fab-7 chromatin boundary. Kyrchanova O, Kurbidaeva A, Sabirov M, Postika N, Wolle D, Aoki T, Maksimenko O, Mogila V, Schedl P, Georgiev P.PLoS Genet. 2018 Aug 15;14(8):e1007442. doi: 10.1371/journal.pgen.1007442. eCollection 2018 Aug.PMID: 30110328
13) Boundaries mediate long-distance interactions between enhancers and promoters in the Drosophila Bithorax complex. Postika N, Metzler M, Affolter M, Müller M, Schedl P, Georgiev P, Kyrchanova O.PLoS Genet. 2018 Dec 12;14(12):e1007702. doi: 10.1371/journal.pgen.1007702. eCollection 2018 Dec.PMID: 30540750
14) Functional dissection of the developmentally restricted BEN domain chromatin boundary factor Insensitive. Fedotova A, Clendinen C, Bonchuk A, Mogila V, Aoki T, Georgiev P, Schedl P.Epigenetics Chromatin. 2019 Jan 3;12(1):2. doi: 10.1186/s13072-018-0249-2.PMID: 30602385
15) The insulator functions of the Drosophila polydactyl C2H2 zinc finger protein CTCF: Necessity versus sufficiency. Kyrchanova O, Maksimenko O, Ibragimov A, Sokolov V, Postika N, Lukyanova M, Schedl P, Georgiev P.Sci Adv. 2020 Mar 25;6(13):eaaz3152. doi: 10.1126/sciadv.aaz3152. eCollection 2020 Mar.PMID: 32232161
16). Boundaries potentiate polycomb response element-mediated silencing. Erokhin M, Gorbenko F, Lomaev D, Mazina MY, Mikhailova A, Garaev AK, Parshikov A, Vorobyeva NE, Georgiev P, Schedl P, Chetverina D.BMC Biol. 2021 Jun 2;19(1):113. doi: 10.1186/s12915-021-01047-8.PMID: 34078365
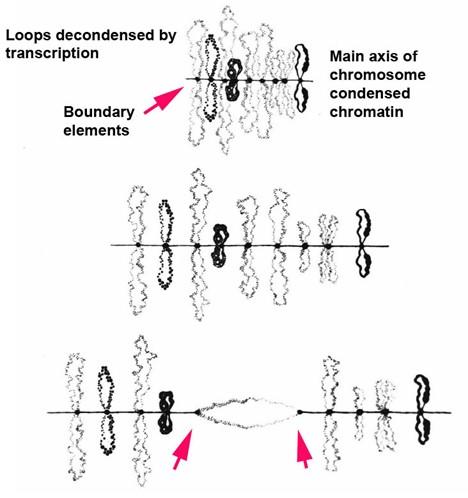
Fig. 1. Lampbrush chrosomoes. When lampbrush chromosomes are stretched pairing interactions between paired boundary elements can be broken. (Callan, 1963).
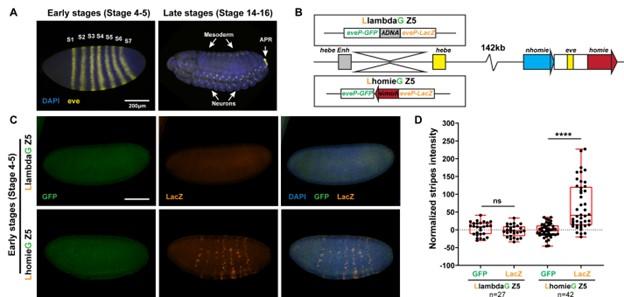
Fig. 2. Pairing interactions between a dual lacZ-GFP reporter (inserted at -142 kb) containing a homie boundary and the homie and nhomie boundaries in the eve-skipped (eve) locus can bring the transgene in close proximity to the eve locus. The eve enhancers can then activate expression of the reporter in early embryos; however, because boundary:boundary pairing interactions are orientation dependent, only one reporter (in this case lacZ) is activated.
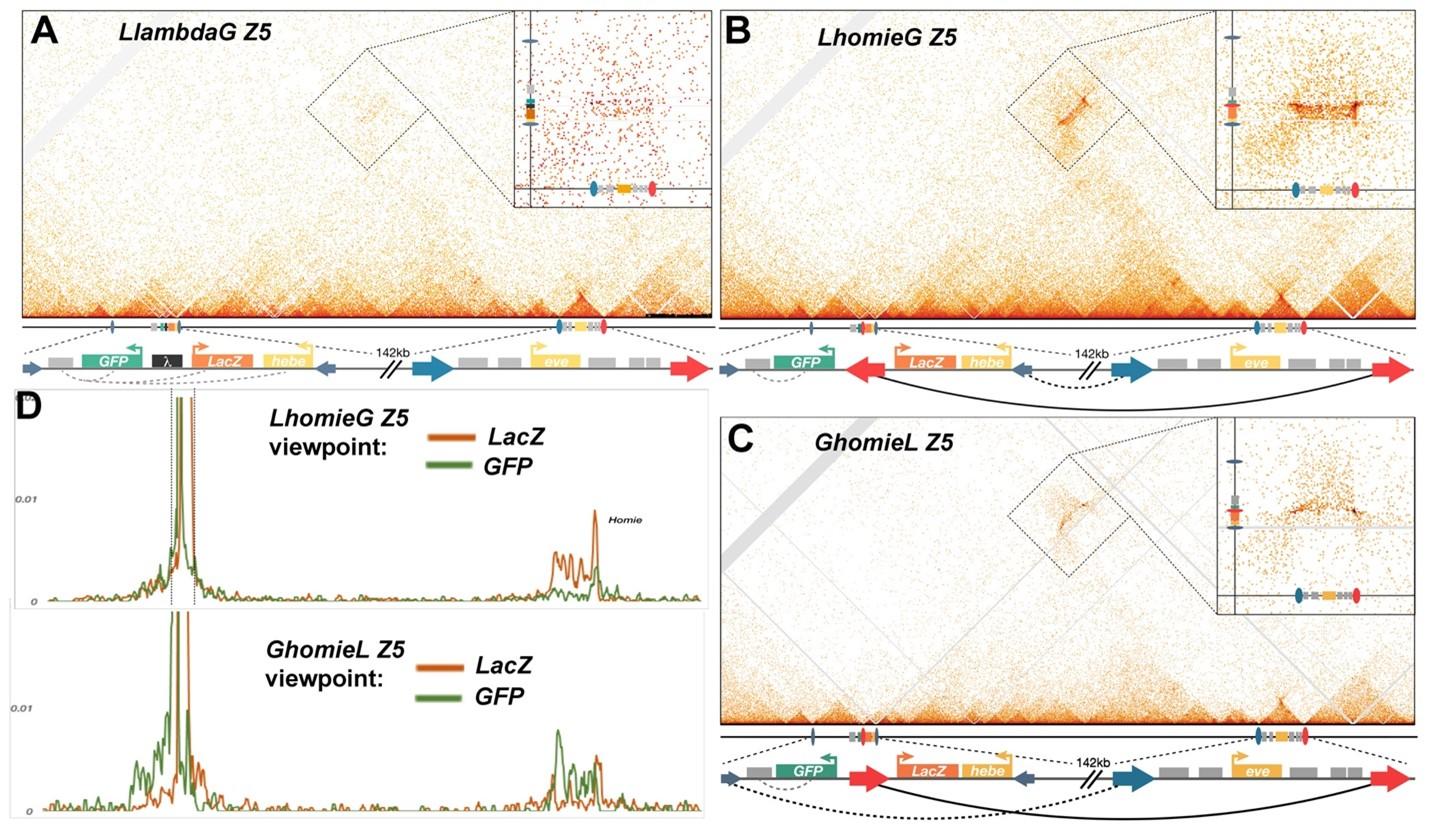
Fig. 3. MicroC of the eve locus and the transgene insertion site at -142 kb. Small dark blue arrows – insulators up and downstream of hebe that appear important to demarcate interaction domains between –142kb and eve. Large blue arrow - endogenous nhomie. Large red arrow – homie, either endogenous or in the transgene. The direction of arrows follow established convention on nhomie/homie, and do not reflect orientation of insulator protein binding motifs per se. Gray boxes – enhancers. A) microC map of the control line LlambdaG Z5. Scaled cartoons of the two loci of interest are shown directly below the microC map, and an unscaled blowup of the elements of interest at each locus is provided. Within the microC map of the entire locus, a zoom-in of the off-diagonal interaction between –142kb insertion cassette and the endogenous eve locus is shown. Note a slight increase in interaction frequency (compare to Figure 1A). B, C) microC map of LhomieG Z5 and GhomieL Z5, respectively. The only difference between the two lines is the orientation of the transgenic homie, as indicate below each Micro-C map. Blow up of the off-diagonal interaction between –142kb and eve, including scaled cartoons denoting features of interest on the top right corner. Note the changed pattern of interaction with endogenous eve locus due to the orientation switch. D) “Virtual 4C” maps obtained from microC maps of “B” on top, and “C” bottom panels, respectively. The viewpoints are shown from either the lacZ gene in orange or the GFP gene in green for both panels. Note the increased interaction of either gene with endogenous eve depending on the change of orientation of homie in each cassette.
Oocyte specification and orb
In many invertebrates and vertebrates, oocytes and sperm are derived from a cyst of cells connected to each other by cytoplasmic bridges. These cysts arise from a single cell, the cystoblast, that undergoes a series of synchronous mitotic divisions with incomplete cytokinesis to give to rise to a multicellular cyst. In testes, all cells typically become sperm. In ovaries, this is also true for some vertebrate species; however, in other species only one member of the cyst becomes an oocyte. In Drosophila five mitotic divisions give rise to a highly invariant 16-cell cyst. These cell divisions occur in the germarium. In the first division, a germline stem cell (GSC) divides to produce two daughters. One daughter remains a GSC, while the other daughter becomes the cystoblast. The cystoblast undergoes four mitotic divisions to generate the 16-cell cyst. Two of the cells in the cyst, the original daughter of the GSC (the cystoblast) and its daughter, have four ring canals. These two cells correspond to the pro-oocytes, and they are linked to each other by a ring canal which was formed at the first division of the cystoblast (Fig. 1). One of the pro-oocytes becomes the oocyte, while the other assumes a nurse cell fate. The pro-oocytes have three additional ring canals, linking the pro-oocytes to their three successive daughters.
Oocyte specification takes place after the formation of the 16-cell cyst and is a robust symmetry breaking process. As the cysts begin to mature in region 2a of the germarium, cytoplasmic markers of oocyte identity and assemble a synaptonemal complex in their nuclei. One of the pro-oocysts is selected to become the oocyte as the 16-cell cysts transition from region 2a to 2b of the germarium. At this point cytoplasmic markers begin accumulating in only one of the pro-oocytes, which then assume an oocyte identity, while the other pro-oocyte loses cytoplasmic markers of oocyte identity, disassembles the synaptonemal complex, and commits to a nurse cell fate.
The available evidence indicates that nurse cell identity is the default fate. Thus, there must be mechanisms in place that make these two pro-oocytes unique amongst all of the cells in the cyst in that they can potentially acquire oocyte identity. In addition, there must mechanisms that drive oocyte fate in one of the two cells, and conversely ensure that the other cell assumes a nurse cell identity.
Key players in oocyte specification include a tubular membranous structure called the fusome which extends through the ring canals connecting all 16 cells in the cysts and two dynein mRNA cargo adaptors, Bicaudal-D (BicD) and Egalitarian (Egl), Mutations in BicD or egl block oocyte specification and in both cases mutant egg chambers have 16 nurse cells and no oocyte The fact that oocyte specification depends upon a dynein adaptor for mRNA cargoes suggests that oocyte specification is driven by mRNAs that are localized to the pro-oocytes. Of the mRNAs that are localize in the pro-oocytes in the germarium, the best candidate for the critical specification factor is orb. orb mRNAs and proteins are amongst the very first gene products that are specifically localized in the pro-oocytes, and subsequently the oocyte. orb encodes one of the two fly CPEB (cytoplasmic polyadenylation element binding protein) translation factors. Proteins in this family bind to hundreds of mRNAs and depending on the biological context can negatively or positively regulate their translation. In addition to being able to control the translation of mRNAs required for oocyte differentiation, orb has a positive autoregulatory activity in which Orb protein binds to the 3’ UTR of orb mRNA and activates its on-site translation (Fig. 2: translation of lacZ-orb 3’ UTR mRNA depends upon orb). In other biological contexts, positive feed-forward loops are used to drive fate specification.
Consistent with the idea that orb mRNAs function as a critical oocyte determinant, when sequences encoding the orb 3’ UTR are deleted (orbD3’UTR) from the endogenous gene, the mutant mRNAs don’t localize to the pro-oocytes, the orb autoregulatory loop (which drives the translation of high levels of Orb protein in the pro-oocytes) is not activated and oocyte specification fails. The orb 3’ UTR is also sufficient to drive oocyte specification when linked to the orb protein coding sequence: The oocyte specification defects of the orbD3’UTR mutant can be partially rescued by introducing a sequence, XN, containing a part of the orb 3’ UTR into the orbD3’UTR deletion. However, the rescuing activity is unusual in that oocyte specification is delayed until stage 2 of oogenesis, which is after the egg chamber exits the germarium. At this point, only half of the chambers successfully specify an oocyte. In these chambers, orb mRNA and protein, as well as other markers of oocyte identity accumulate in a single cell and this cell develops as the oocyte. As is the case in wild type, the oocyte is always one of the two cells that have four ring canals. In the chambers that failed to specify an oocyte, orb mRNAs and proteins are distributed throughout in all 16 germ cells as are other markers of oocyte identity, and all cells develop as nurse cells.
Ongoing studies are focused on the mechanisms that localize and then anchor orb mRNA in the two pro-oocytes, and on the activation of the orb autoregulatory loop so that high levels of protein accumulate in these two cells. We find that a critical step for specification of the oocyte in the germarium is the tight association of orb mRNA with the fusome. orb mRNA association with the fusome requires the orb 3’ UTR and Orb protein (Fig. 3: orb 3’ UTR mediates fusome association). When orb mRNAs are not linked to the fusome in region 2a/2b, oocyte specification fails. This fusome dependence is bypassed in XN; however, oocyte specification doesn’t take place till after the egg chambers exit the germarium.
Publications:
1) The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P.Genes Dev. 1994 Mar 1;8(5):598-613. doi: 10.1101/gad.8.5.598.PMID: 7523244
2) An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Tan L, Chang JS, Costa A, Schedl P.Development. 2001 Apr;128(7):1159-69. doi: 10.1242/dev.128.7.1159.PMID: 11245581
3) The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA.Dev Cell. 2005 Mar;8(3):331-42. doi: 10.1016/j.devcel.2005.01.011.PMID: 15737929
4) Cup blocks the precocious activation of the orb autoregulatory loop. Wong LC, Schedl P.PLoS One. 2011;6(12):e28261. doi: 10.1371/journal.pone.0028261. Epub 2011 Dec 2.PMID: 22164257
5). The Drosophila CPEB Protein Orb Specifies Oocyte Fate by a 3'UTR-Dependent Autoregulatory Loop. Barr J, Gilmutdinov R, Wang L, Shidlovskii Y, Schedl P.Genetics. 2019 Dec;213(4):1431-1446. doi: 10.1534/genetics.119.302687. Epub 2019 Oct 8.PMID: 31594794
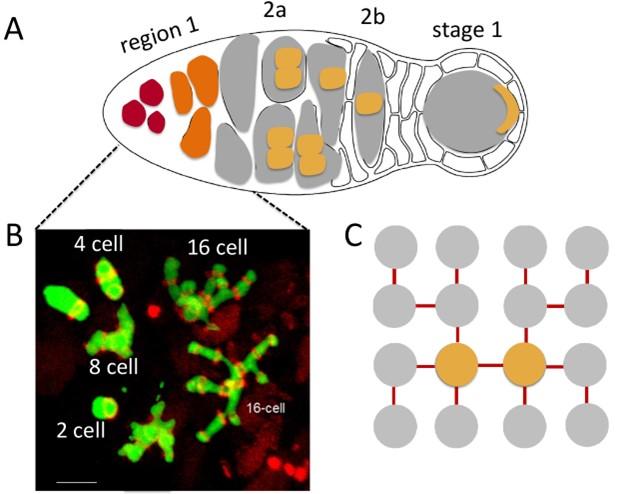
Fig. 1 Formation of the 16 cell cysts plus fusome organization during the mitotic divisions. (orange: pro-oocytes.
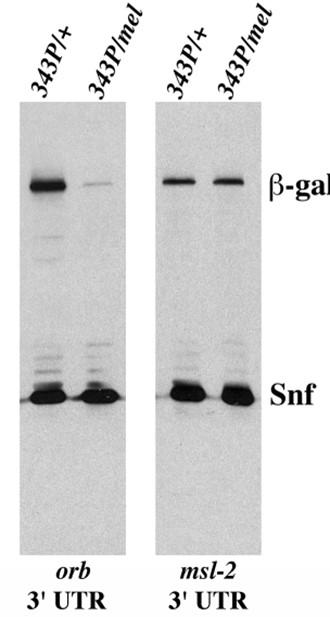
Fig. 2. Orb protein binds to orb 3’ UTR and regulates its own synthesis. Expression of LacZ in hsp83 promoter: lacZ orb 3’ UTR and hsp83promoter:lacZ msl-2 3’UTR in orb343/+ and orb343/ orbmel female ovaries. orb343 is a null allele, while orbmel is a weak hypomorph.
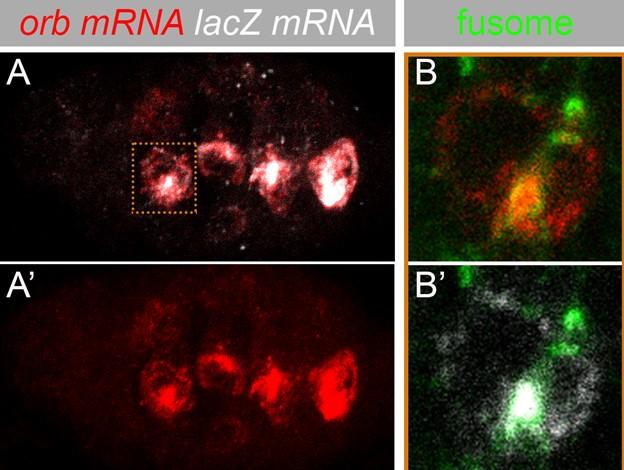
Fig. 3. Association of orb mRNA and a chimeric lacZ orb 3’ UTR with the fusome in regions 2a/2b of the germarium.
Formation of the Embryonic Gonad
PGC migration
In Drosophila melanogaster, the embryonic gonad is made up of two cell types, the primordial germ cells (PGCs) and the somatic gonadal precursor cells (SGPs). As the PGCs and SGPs are specified at distant sites, gonad formation requires directed migration, recognition and sustained association between these two cell types. Specification of the SGPs, which are mesodermal in origin occurs during mid-embryogenesis, and depends on the action of zygotic patterning determinants. In contrast, PGCs contrast, are specified earlier at the blastoderm stage, on the external surface of the embryo, and are formed under the control of maternal determinants located at the posterior pole of the egg during oogenesis and the BMP signaling pathway in the zygote. The PGCs must find their way from their site of formation on the outside surface of the embryo at the posterior pole to the mesodermally derived somatic gonadal precursor cells (SGPs) to form the primitive embryonic gonad. The migratory journey is initiated during gastrulation when the PGCs are brought into the interior of the embryo by the midgut invagination. They must then pass through the midgut and make their way through the mesoderm to reach the SGPs, which are located in parasegments (PS) 10-13. After the PGCs and SGP come into contact, they first align with each other across several parasegments and them coalesce into the embryonic gonad so that the PGCs are on the inside and the SGPs are on the outside, they line up with each other along the mesoderm.
This migratory journey is orchestrated by a novel ‘non-canonical’ hedgehog (hh) signaling pathway which inveigles the PGCs to migrate towards the SGPs (Fig.1:ectopic hh induces mismigration). The attractive cues provided by hh are complemented by a repulsive “signal” that helps guide the migrating PGCs at critical junctures in the appropriate directions. The repulsive signal is generated by two lipid phosphate phosphatases, wunen and wunen-2.
The deployment of a classical morphogen as a guidance molecule poses many interesting questions. One of these is the mechanism of signal transduction. Like other signaling pathways, hh is known to signal over long distances. Moreover, at this stage of development there are many sources of Hh not only in the ectoderm, but also in the mesoderm parasegments anterior to PS10. What distinguishes the Hh ligand directing PGC migration from Hh ligands that are responsible for non-autonomous fate specification? One mechanism is the potentiation of hh signals from the SGPs mediated by hmgcr. hmgcr is required for the transmission of the Hh lingand (Fig. 2). In the classical view the SGPs are thought to secrete the guidance molecules into the extracellular space generating a diffusion gradient which inveigles the PGCs to migrate towards the source. However, long distance signaling by the Hh pathway is known to be mediated by special cytoplasmic extensions called cytonemes. Cytonemes containing the Hh ligand extend from hh expressing cells and are met by cytonemes containing the Patched receptor that extend from hh receiving cells. Is this the mechanism that is used to guide the migrating PGCs? In the canonical hh signaling pathway, reception of the Hh signal by the receiving cell induces a signaling cascade that results in transcriptional response. However, directed cell migration requires modulating the function of the cytoskeleton, not transcriptional activation. What the nature of the non- canonical pathway that remodels the cytoskeleton so that PGCs can move towards the SGPs (Fig. 3)?
Selected Publications:
1) hedgehog signaling in germ cell migration. Deshpande G, Swanhart L, Chiang P, Schedl P.Cell. 2001 Sep 21;106(6):759-69. doi: 10.1016/s0092-8674(01)00488-3.PMID: 11572781
2) HMGCoA reductase potentiates hedgehog signaling in Drosophila melanogaster. Deshpande G, Schedl P.Dev Cell. 2005 Nov;9(5):629-38. doi: 10.1016/j.devcel.2005.09.014. PMID: 16256738
3) The hedgehog pathway gene shifted functions together with the hmgcr-dependent isoprenoid biosynthetic pathway to orchestrate germ cell migration. Deshpande G, Zhou K, Wan JY, Friedrich J, Jourjine N, Smith D, Schedl P.PLoS Genet. 2013;9(9):e1003720. doi: 10.1371/journal.pgen.1003720. Epub 2013 Sep 12.PMID: 24068944
4) Role of the ABC transporter Mdr49 in Hedgehog signaling and germ cell migration. Deshpande G, Manry D, Jourjine N, Mogila V, Mozes H, Bialistoky T, Gerlitz O, Schedl P.Development. 2016 Jun 15;143(12):2111-20. doi: 10.1242/dev.133587. Epub 2016 Apr 27.PMID: 27122170
Fig. 1 Ectopic expression of Hh disrupts PGC migration
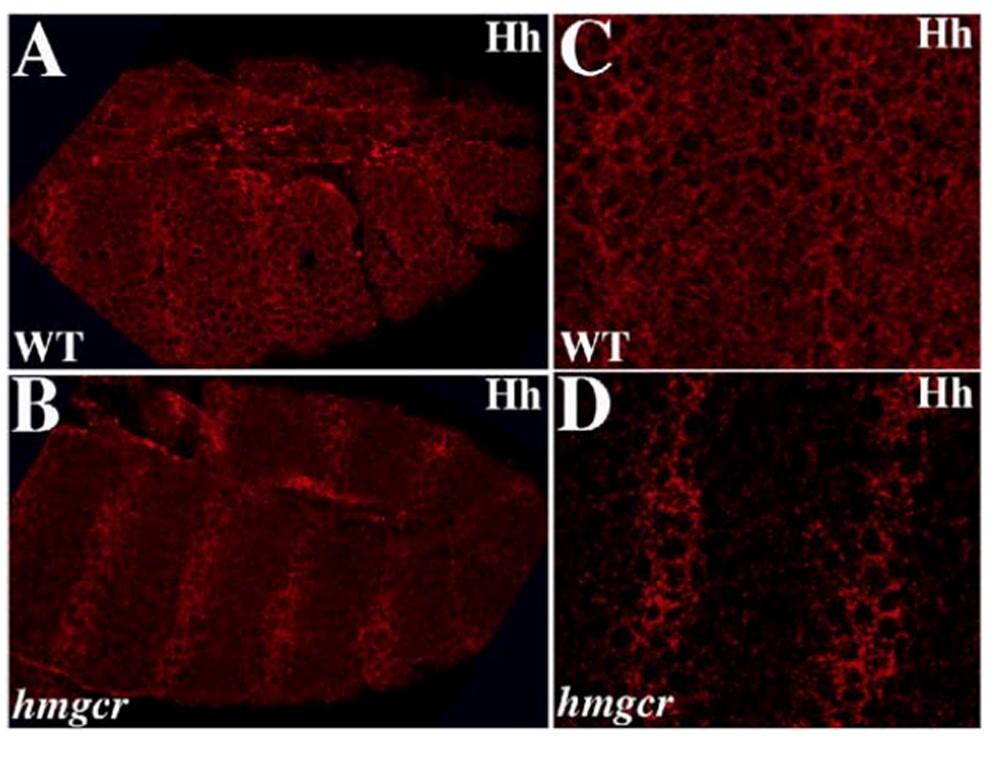
Fig. 2 hmgcr is required for releasing from hh expressing cells.

Fig. 3 Ca2+ influxes in migrating PGCs. Frame every 6 sec.
PGC fate specification
In flies, PGCs are formed after the onset of the minor wave of ZGA, which begins at nuclear cycle (NC) 8. Like many other organisms, transcription is downregulated when the PGCs are specified. Ongoing transcription is turned off by germ-cell less (gcl) while genes activated during the major wave of ZGA (NC14) are kept off by combined action of polar granule component (pgc) and nanos (nos) (Fig. 4). Broad downregulation of transcription is reflected in the phosphorylation status of the RNA polymerase II CTD domain and the absence of H3meK4, a modification associated with active transcription. Another feature common to worms, flies and mammals, PGCs arrest cell cycle in G2. Moreover, genes implicated in PGC specification in model invertebrates (flies or worms), like nos, vasa, and piwi, are also conserved in higher animals and appear to have a similar germline function. While many of the characteristics of PGCs that distinguish them from the soma are widely shared amongst different animal species, there is one striking dichotomy, namely whether the mechanism driving specification is “epigenesis” or “preformation.” In epigenesis, specification is non-autonomous and depends upon cell-cell signaling. In preformation, specification is autonomous and is driven by determinants that are localized in the presumptive PGCs. Mammals utilize epigenesis. In pre-implantation embryos, a combination of inductive Wingless (Wg) and Bone morphogenetic protein (BMP) signals from the extra embryonic ectoderm and visceral endoderm acts to induce cells within the posterior epiblast to become PGCs. By contrast, PGC specification in flies is the classic example of an exclusively preformation. Cell-autonomous factors localized to the posterior pole of the egg by an oskar dependent mechanism during the late stages of oogenesis are thought to be both necessary and sufficient for PGC specification in early embryos. During pole bud formation, the centrosomes/microtubule network associated with each incoming nucleus triggers the release of the localized PGC determinants from the posterior cortical cytoskeleton, and these factors are then incorporated into the newly formed PGCs during cellularization (Fig. 5). When these factors are not properly sequestered in the newly formed PGCs, PGC specification fails. While the importance of maternally localized determinants has known for decades, recent studies have shown that PGC specification in flies is not exclusively preformation as has long been thought. Instead, BMP signaling from the soma functions in conjunction with the oskar localized maternal determinants to specify PGC fate (Fig 6).
Selected Publications
1) Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Deshpande G, Calhoun G, Yanowitz JL, Schedl PD.Cell. 1999 Oct 29;99(3):271-81. doi: 10.1016/s0092-8674(00)81658-x.PMID: 10555143
2) Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Deshpande G, Calhoun G, Schedl P.Development. 2004 Mar;131(6):1247-57. doi: 10.1242/dev.01004. Epub 2004 Feb 11.PMID: 14960492
3) Germ Cell-less Promotes Centrosome Segregation to Induce Germ Cell Formation. Lerit DA, Shebelut CW, Lawlor KJ, Rusan NM, Gavis ER, Schedl P, Deshpande G.Cell Rep. 2017 Jan 24;18(4):831-839. doi: 10.1016/j.celrep.2016.12.074.PMID: 28122234
4) Antagonism between germ cell-less and Torso receptor regulates transcriptional quiescence underlying germline/soma distinction. Colonnetta MM, Lym LR, Wilkins L, Kappes G, Castro EA, Ryder PV, Schedl P, Lerit DA, Deshpande G.Elife. 2021 Jan 18;10:e54346. doi: 10.7554/eLife.54346.PMID: 33459591
5) Preformation and epigenesis converge to specify primordial germ cell fate in the early Drosophila embryo. Colonnetta MM, Goyal Y, Johnson HE, Syal S, Schedl P, Deshpande G.PLoS Genet. 2022 Jan 5;18(1):e1010002. doi: 10.1371/journal.pgen.1010002. eCollection 2022 Jan.PMID: 34986144
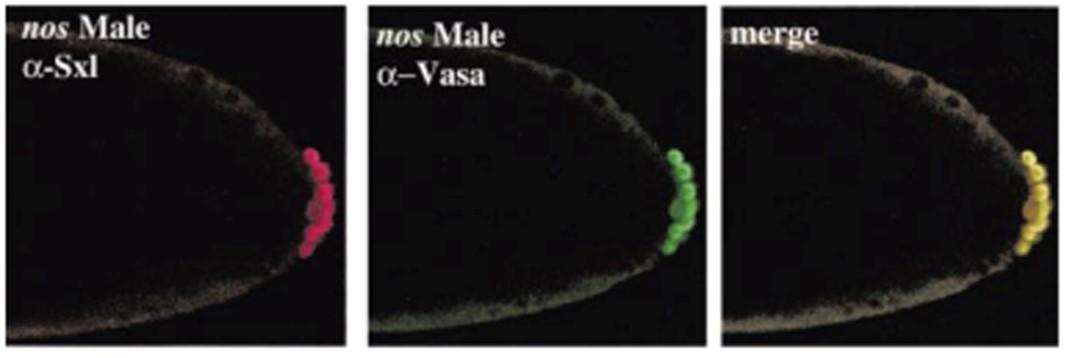
Fig. 4 Induction of the female specific Sex-lethal gene in nos mutant male PGCs
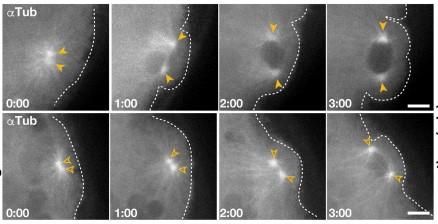
Fig. 5. Centrosome defects in gcl embryos during PGC cellularization (pole bud stage).
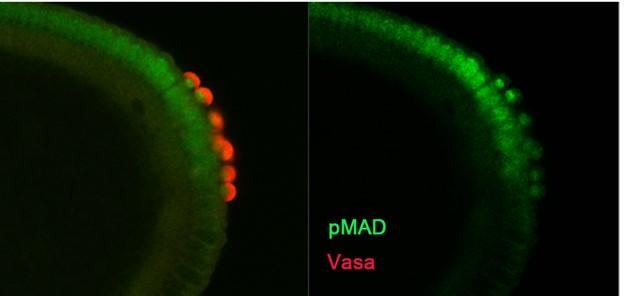
Fig 6 Nuclear localization of the BMP transcription factor pMAD in newly formed PGC nuclei induced by somatic BMP signals.
Documents from 2001/2 “Controversy”
2 Report to Cell by Dr. Dinardo
3 Report to Cell by Deshpande et al.
4 Manuscript Sept. 2022: The Hedgehog pathway and germ cell migration in Drosophila: Res Ipsa Loquitur

